Research
Understanding by Building
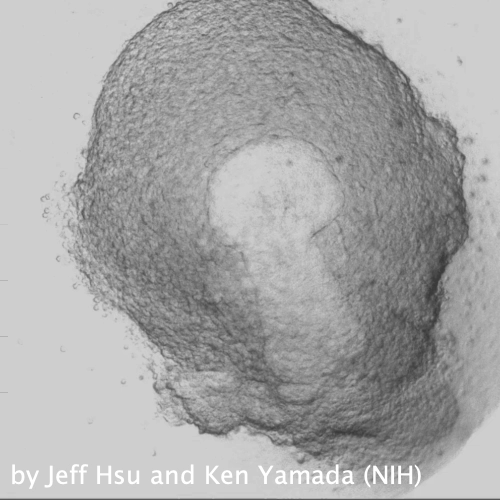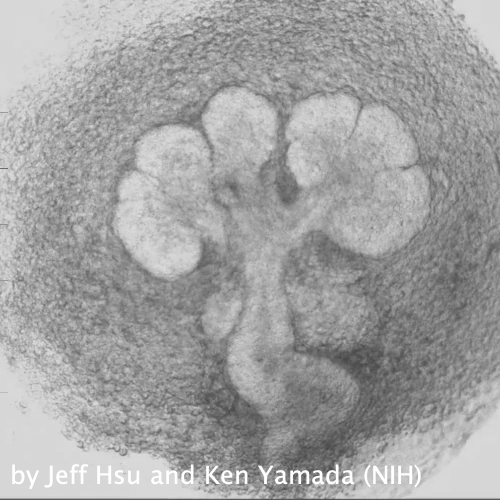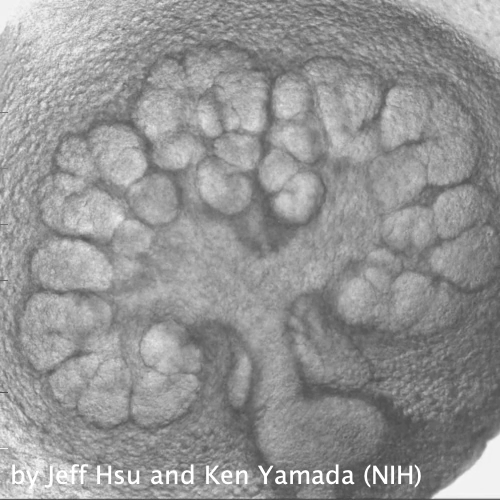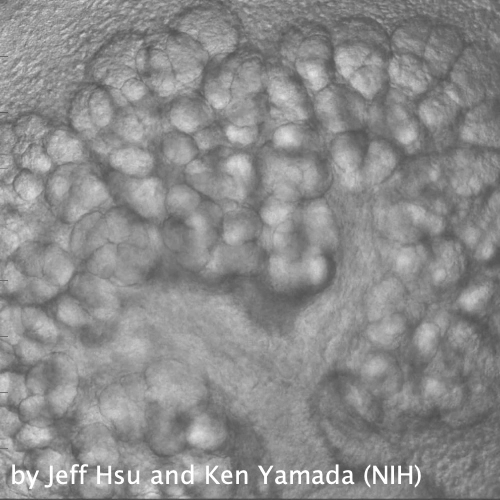
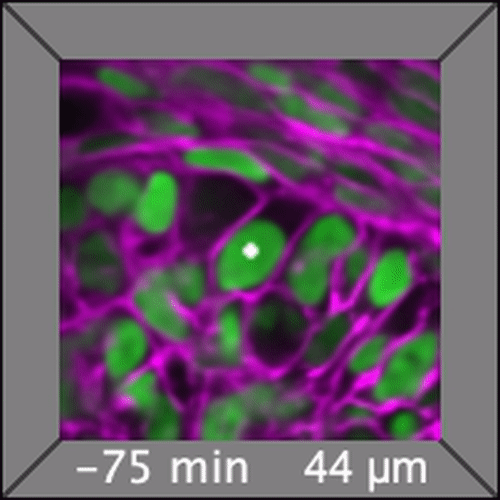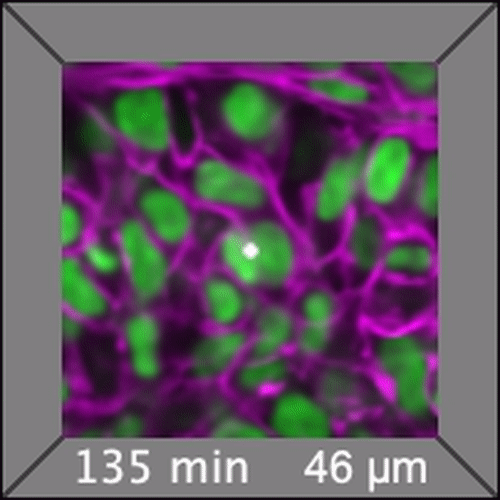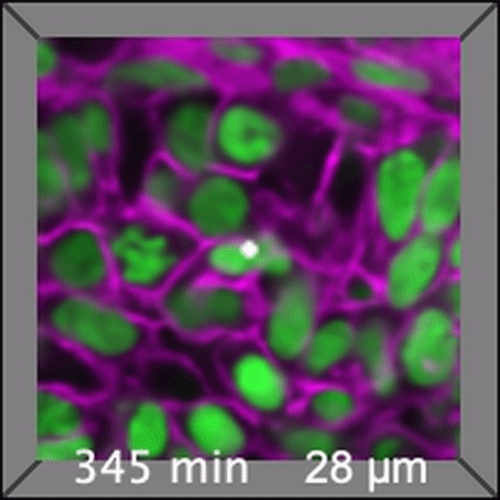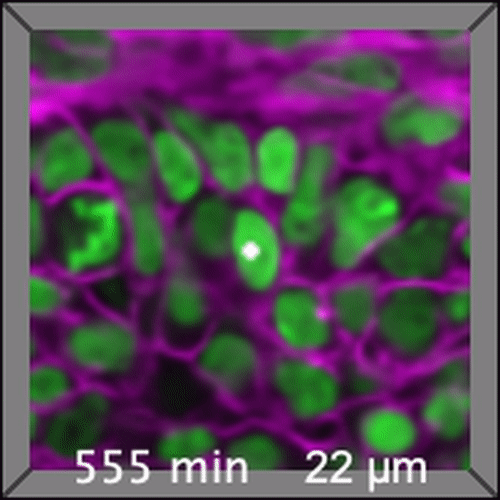
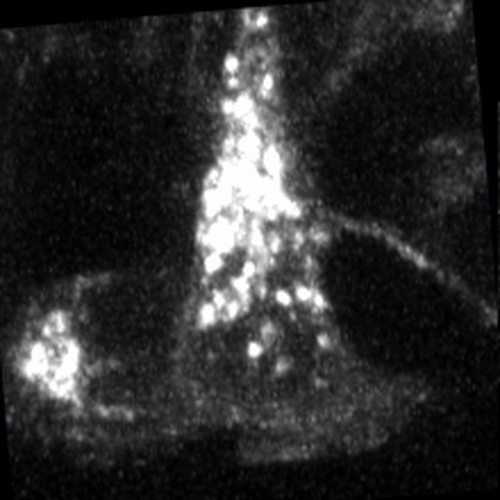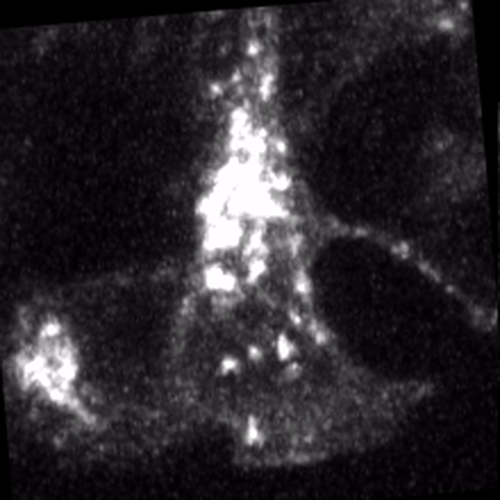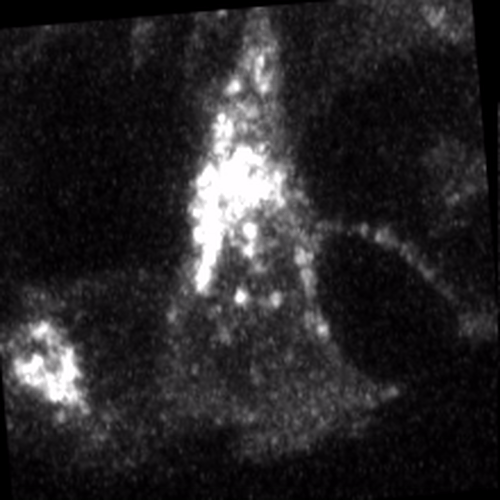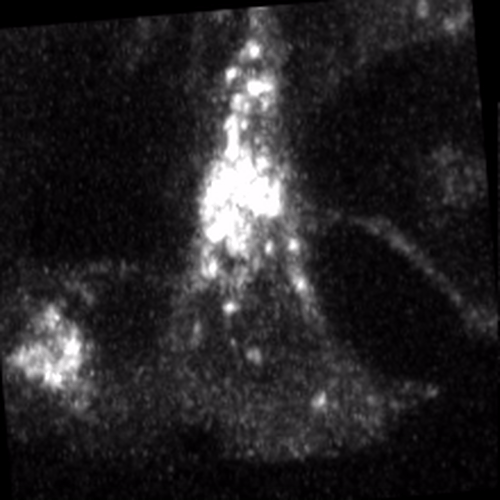
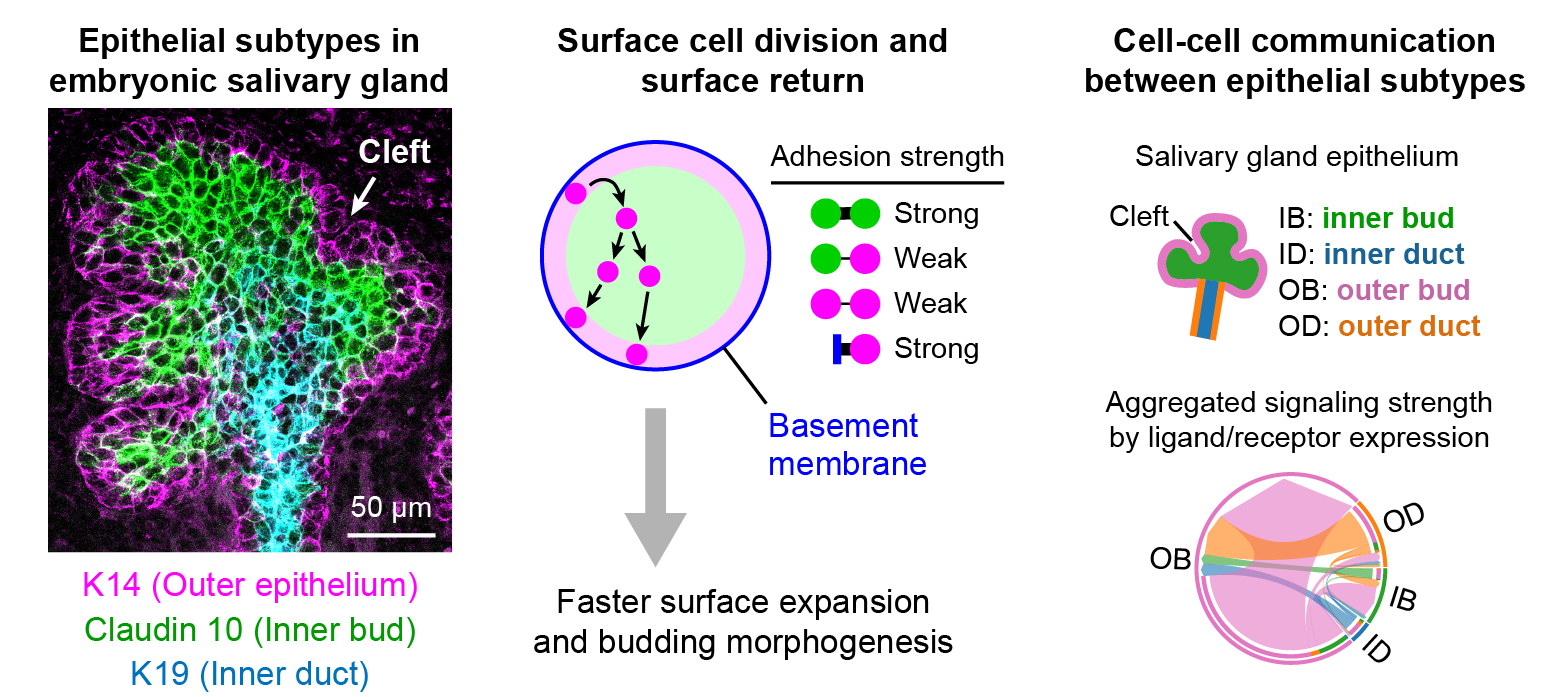
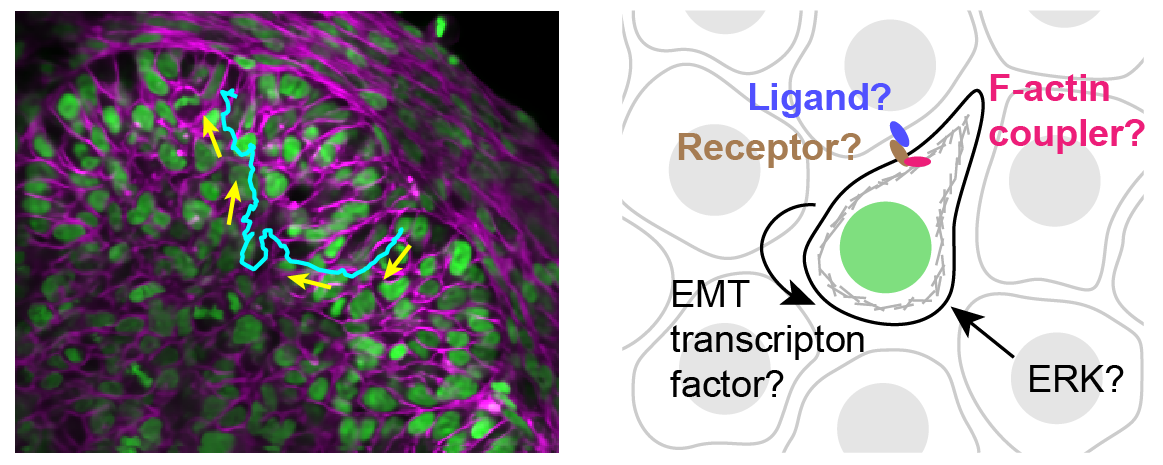
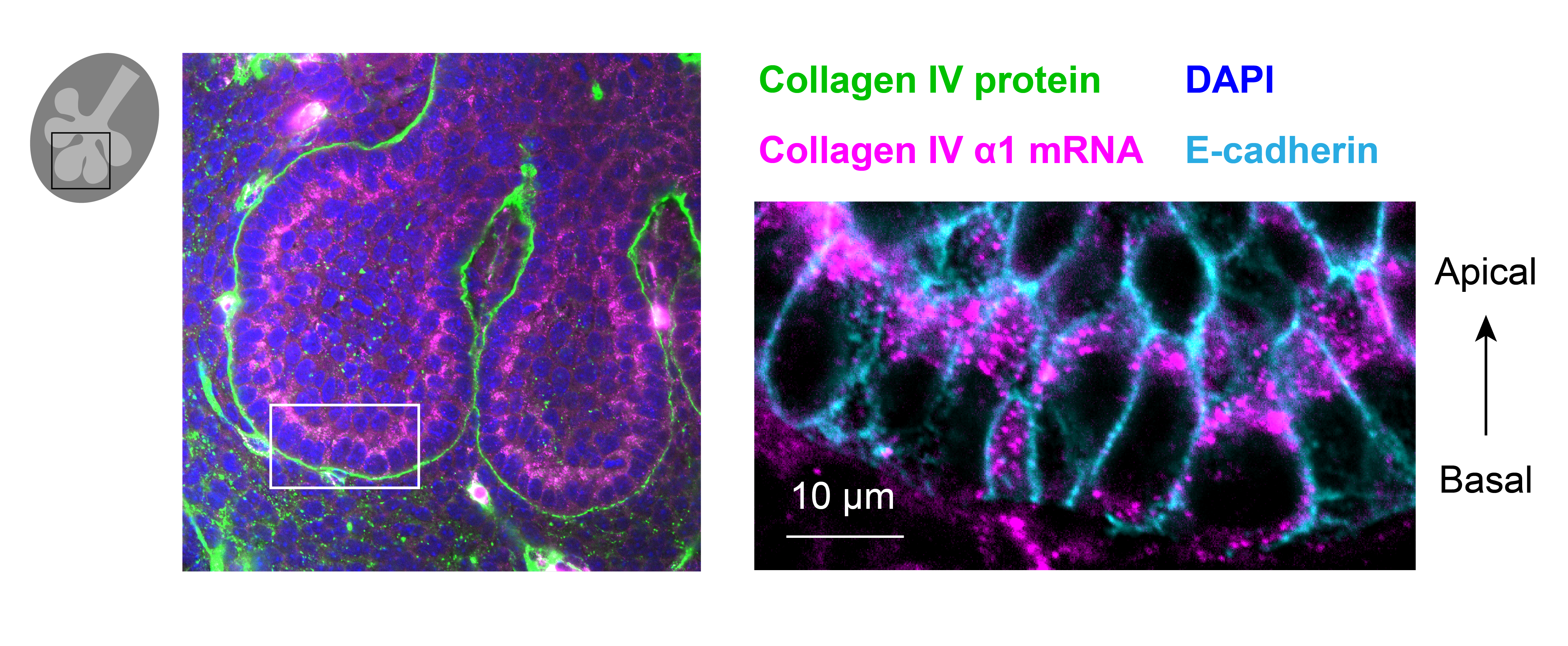
Through coordinated tissue morphogenesis and cell specialization, organ development generates complex structures tailored for diverse physiological functions. For example, lungs, kidneys, and various glands generate tree-like branched structures, where numerous end buds specialized for air exchange, blood filtration, or secretion are connected by a hierarchical ductal system. The mature branched structure arises from a single epithelial bud at the embryonic stage through branching morphogenesis, which requires the dynamic interplay between cells and the extracellular matrix. We are only beginning to understand the principles of branching morphogenesis and other processes in organ development, which will be crucial for developing gene, cell, and tissue engineering-based therapies for defective organs.
We combine embryonic and reconstitution approaches to discover principles of organ development. We employ live imaging, sequencing, spatial transcriptomics, genetic perturbation, and targeted protein degradation to probe native organ development, primarily (but not limited to) using the mouse salivary gland as a versatile model. Driven by the “understanding by building” philosophy, we always try to reconstitute key biological processes from neutral building blocks. Whenever applicable, we use mathematical or physical modeling to theorize our findings. We aspire to pave the way of building customizable organs from the bottom up.
Click on the movies to learn more: